Header file for gensvm_cv_util.c. More...
#include "gensvm_base.h"
Go to the source code of this file.
Functions | |
| void | gensvm_make_cv_split (long N, long folds, long *cv_idx) |
| Create a cross validation split vector. More... | |
| void | gensvm_get_tt_split (struct GenData *full_data, struct GenData *train_data, struct GenData *test_data, long *cv_idx, long fold_idx) |
| Wrapper around sparse/dense versions of this function. More... | |
| void | gensvm_get_tt_split_dense (struct GenData *full_data, struct GenData *train_data, struct GenData *test_data, long *cv_idx, long fold_idx) |
| Create train and test datasets for a CV split with dense data. More... | |
| void | gensvm_get_tt_split_sparse (struct GenData *full_data, struct GenData *train_data, struct GenData *test_data, long *cv_idx, long fold_idx) |
| Create train and test dataset for a CV split with sparse data. More... | |
Detailed Description
Header file for gensvm_cv_util.c.
- Date
- 2014-01-07
Contains function declarations for functions needed for performing cross validation on GenData structures.
- Copyright
- Copyright 2016, G.J.J. van den Burg.
This file is part of GenSVM.
GenSVM is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
GenSVM is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with GenSVM. If not, see http://www.gnu.org/licenses/.
Definition in file gensvm_cv_util.h.
Function Documentation
◆ gensvm_get_tt_split()
| void gensvm_get_tt_split | ( | struct GenData * | full_data, |
| struct GenData * | train_data, | ||
| struct GenData * | test_data, | ||
| long * | cv_idx, | ||
| long | fold_idx | ||
| ) |
Wrapper around sparse/dense versions of this function.
This function tests if the data in the full_data structure is stored in a dense matrix format or not, and calls gensvm_get_tt_split_dense() or gensvm_get_tt_split_sparse() accordingly.
- Parameters
-
[in] full_data a GenData structure for the entire dataset [in,out] train_data an initialized GenData structure which on exit contains the training dataset [in,out] test_data an initialized GenData structure which on exit contains the test dataset [in] cv_idx a vector of cv partitions created by gensvm_make_cv_split() [in] fold_idx index of the fold which becomes the test dataset
Definition at line 107 of file gensvm_cv_util.c.

◆ gensvm_get_tt_split_dense()
| void gensvm_get_tt_split_dense | ( | struct GenData * | full_data, |
| struct GenData * | train_data, | ||
| struct GenData * | test_data, | ||
| long * | cv_idx, | ||
| long | fold_idx | ||
| ) |
Create train and test datasets for a CV split with dense data.
Given a GenData structure for the full dataset, a previously created cross validation split vector and a fold index, a training and test dataset are created. It is assumed here that the data is stored as a dense matrix, and that the train and test data should also be stored as a dense matrix.
- Parameters
-
[in] full_data a GenData structure for the entire dataset [in,out] train_data an initialized GenData structure which on exit contains the training dataset [in,out] test_data an initialized GenData structure which on exit contains the test dataset [in] cv_idx a vector of cv partitions created by gensvm_make_cv_split() [in] fold_idx index of the fold which becomes the test dataset
Definition at line 142 of file gensvm_cv_util.c.
◆ gensvm_get_tt_split_sparse()
| void gensvm_get_tt_split_sparse | ( | struct GenData * | full_data, |
| struct GenData * | train_data, | ||
| struct GenData * | test_data, | ||
| long * | cv_idx, | ||
| long | fold_idx | ||
| ) |
Create train and test dataset for a CV split with sparse data.
Given a GenData structure for the full dataset, a previously created cross validation split vector and a fold index, a training and test dataset are created. It is assumed here that the data is stored as a sparse matrix, and that the train and test data should also be stored as a sparse matrix.
- Parameters
-
[in] full_data a GenData structure for the entire dataset [in,out] train_data an initialized GenData structure which on exit contains the training dataset [in,out] test_data an initialized GenData structure which on exit contains the test dataset [in] cv_idx a vector of cv partitions created by gensvm_make_cv_split() [in] fold_idx index of the fold which becomes the test dataset
Definition at line 223 of file gensvm_cv_util.c.
◆ gensvm_make_cv_split()
| void gensvm_make_cv_split | ( | long | N, |
| long | folds, | ||
| long * | cv_idx | ||
| ) |
Create a cross validation split vector.
A pre-allocated vector of length N is created which can be used to define cross validation splits. The folds are contain between 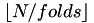 and
and 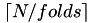 instances. An instance is mapped to a partition randomly until all folds contain
instances. An instance is mapped to a partition randomly until all folds contain 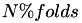 instances. The zero fold then contains
instances. The zero fold then contains 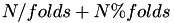 instances. These remaining
instances. These remaining 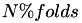 instances are then distributed over the first
instances are then distributed over the first 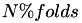 folds.
folds.
- Parameters
-
[in] N number of instances [in] folds number of folds [in,out] cv_idx array of size N which contains the fold index for each observation on exit
Definition at line 54 of file gensvm_cv_util.c.
 1.8.13
1.8.13