This page specifies the training file that can be parsed by read_training_from_file(). Below is an example training file.
train: /path/to/training/dataset.txt test: /path/to/test/dataset.txt p: 1.0 1.5 2.0 kappa: -0.9 0.0 1.0 lambda: 64 16 4 1 0.25 0.0625 0.015625 0.00390625 0.0009765625 0.000244140625 epsilon: 1e-6 weight: 1 2 folds: 10 kernel: LINEAR gamma: 1e-3 1e-1 1e1 1e3 coef: 1.0 2.0 degree: 2.0 3.0
Note that with a LINEAR kernel specification, the gamma, coef, and degree parameters do not need to be specified. The above merely shows all available parameters that can be specified in the grid search. Below each of the parameters are described in more detail. Arguments followed by an asterisk are optional.
train:
The location of the training dataset file. See Default Data File Specification for the specification of a dataset file.
test:*
The location of a test dataset file. See Default Data File Specification for the specification of a dataset file. This is optional, if specified the train/test split will be used for training.
p:
The values of the p parameter of the algorithm to search over. The p parameter is used in the 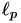 norm over the Huber weighted scalar misclassification errors. Note:
norm over the Huber weighted scalar misclassification errors. Note: 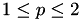 .
.
kappa:
The values of the kappa parameter of the algorithm to search over. The kappa parameter is used in the Huber hinge error over the scalar misclassification errors. Note: 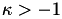 .
.
lambda:
The values of the lambda parameter of the algorithm to search over. The lambda parameter is used in the regularization term of the loss function. Note: 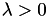 .
.
epsilon:
The values of the epsilon parameter of the algorithm to search over. The epsilon parameter is used as the stopping parameter in the majorization algorithm. Note that it often suffices to use only one epsilon value. Using more than one value increases the size of the grid search considerably.
weight:
The weight specifications for the algorithm to use. Two weight specifications are implemented: the unit weights (index = 1) and the group size correction weights (index = 2). See also gensvm_initialize_weights().
folds:
The number of cross validation folds to use.
kernel:*
Kernel to use in training. Only one kernel can be specified. See KernelType for available kernel functions. Note: if multiple kernel types are specified on this line, only the last value will be used (see the implementation of parse_kernel_str() for details). If no kernel is specified, the LINEAR kernel will be used.
gamma:*
Gamma parameters for the RBF, POLY, and SIGMOID kernels. This parameter is only optional if the LINEAR kernel is specified. See gensvm_kernel_dot_rbf(), gensvm_kernel_dot_poly(), and gensvm_kernel_dot_sigmoid() for kernel specifications.
coef:*
Coefficients for the POLY and SIGMOID kernels. This parameter is only optional if the LINEAR or RBF kernels are used. See gensvm_kernel_dot_poly(), and gensvm_kernel_dot_sigmoid() for kernel specifications.
degree:*
Degrees to search over in the grid search when the POLY kernel is specified. With other kernel specifications this parameter is unnecessary. See gensvm_kernel_dot_poly() for the polynomial kernel specification.
 1.8.13
1.8.13